Peak fitting XRD data with Python
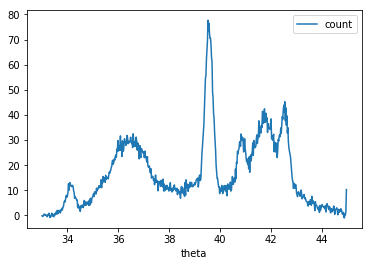
While it may not be apparent on my blog, I am graduate student studying computational material science. Our group studies the fundamental physics behind ion beam modification and radiation resistant nuclear materials. One of the techniques our experimentalists use regularly is x-ray diffraction (XRD). XRD is a technique where you point an x-ray beam at a material in a set angle and observe the resulting angles and intensities of the diffracted beam. These patterns when measures with a camera along all angles $\theta$ result in peaks. The figure below shows an example pattern of ours.
It turns out that all XRD profiles are a combination of gaussians, lorentzians, and voigt functions. As a material scientist I had always been told that fitting is hard. But the mathematician side of me never believed it! There are numerous commercially available and open source softwares to do this job such as Origin Pro Peak Fitting, IGOR Pro, GSAS, and many others. But to me they all have one problem. They obscure the simple mathematics taking place behind the scenes. Infact in this post I will show how with numpy and scipy alone we can create our own peak fitting software that is just as successful. Later we will use the excellent python package lmfit which automates all the tedious parts of writting our own fitting software. First lets import everything we will need.
|
|
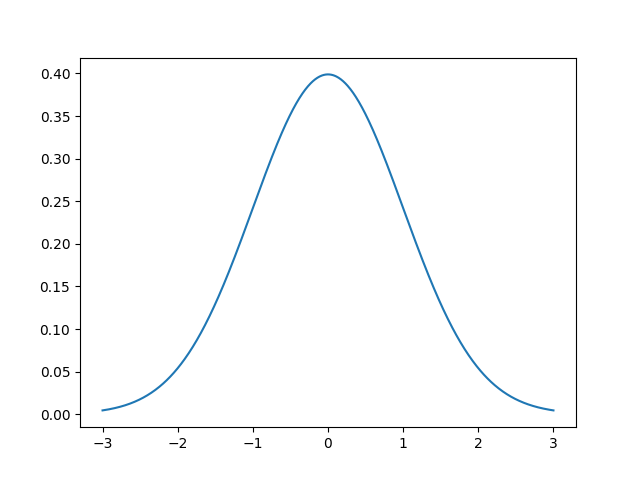
Now I promise we will get to fitting this XRD profile but first we must show what is involved in fitting gaussians, lorentzians, and voigt functions. In general this fitting process can be written as non-linear optimization where we are taking a sum of functions to reproduce the data. The gaussian function is also known as a normal distribution. In the figure below we show a gaussian with amplitude 1, mean 0, and variance 1.
$$f(x; A, \mu, \sigma) = \frac{A}{\sigma\sqrt{2\pi}} e^{[{-{(x-\mu)^2}/{{2\sigma}^2}}]}$$
|
|
Now I will show simple optimization using scipy which we will use
for solving for this non-linear sum of functions. There is a really
nice scipy.optimize.minimize method that has several optimizers. I
will only use the default one for these demonstrations. Let’s see this
method in action. Suppose we have an unknown function plotted bellow.
|
|
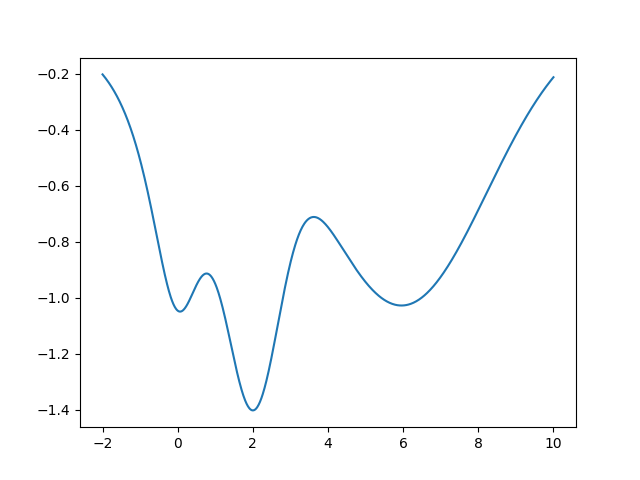
Now we will use scipy.optimize.minimize to find the global minimum
of the function.
|
|
| initial | iterations | minimum |
|---|---|---|
| -0.5 | 5 | 0.064 |
| -2.0 | 4 | 2.001 |
| +9.0 | 3 | 5.955 |
Notice how different initial guesses result in different minimums. This is true for any global optimization problem. These optimization routines do not guarantee that they have found the global minimum. A common issue we will see with fitting XRD data is that there are many of these local minimums where the routine gets stuck. This is why a good initial guess is extremely important. Let’s use this optimization to fit a gaussian with some noise. See the plot below for the data we are trying to fit.
A = 100.0 # intensity
μ = 4.0 # mean
σ = 4.0 # peak width
n = 200
x = np.linspace(-10, 10, n)
y = g(x, A, μ, σ) + np.random.randn(n)
fig, ax = plt.subplots()
ax.scatter(x, y, s=2)
plot_to_blog(fig, 'xrd-fitting-gaussian-noise.png')
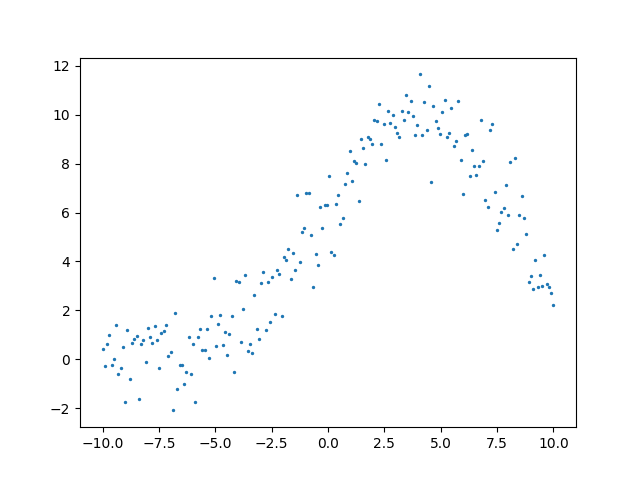
Now we will use optimization to backout what it believes the parameters of the gaussian are. In order to optimize the algorithm needs some way to measure improvement. For this we will use the mean square error as a cost function for our optimization routine.
$$MSE = \frac{1}{n} \sum^n_{i=1} (Y_i - \hat{Y}_i)^2$$
Here $Y_i$ and $\hat{Y}_i$ represents the model values and observed values respectively. The better the fit the closer the MSE will be to zero.
|
|
steps 15 1.0846693928147109
amplitude: 100.103 mean: 3.951 sigma: 3.980
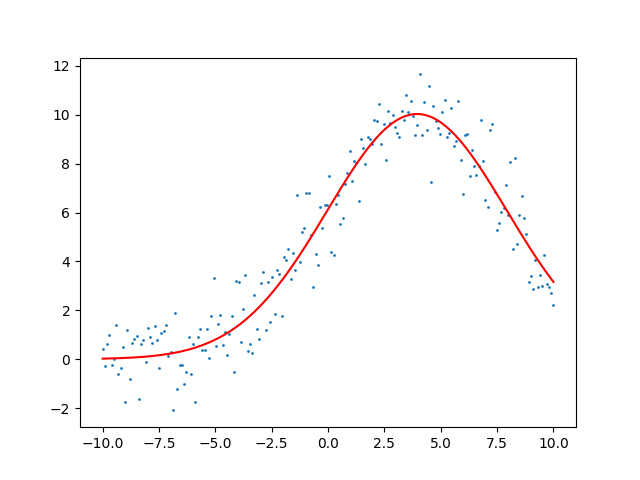
That’s a great fit for some simple data lets throw a harder problem at it. Let’s fit two Gaussian.
|
|
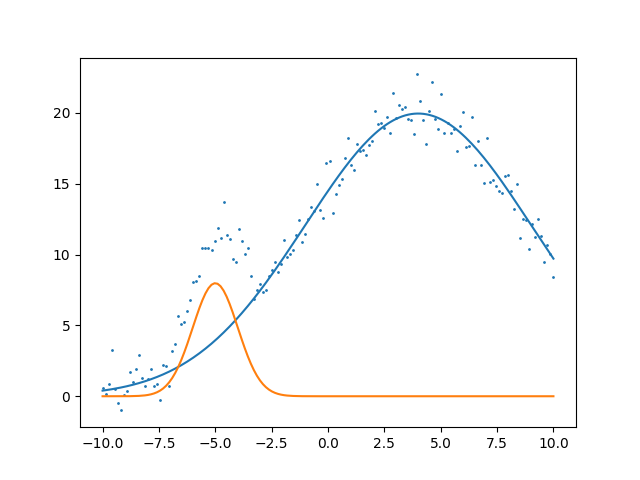
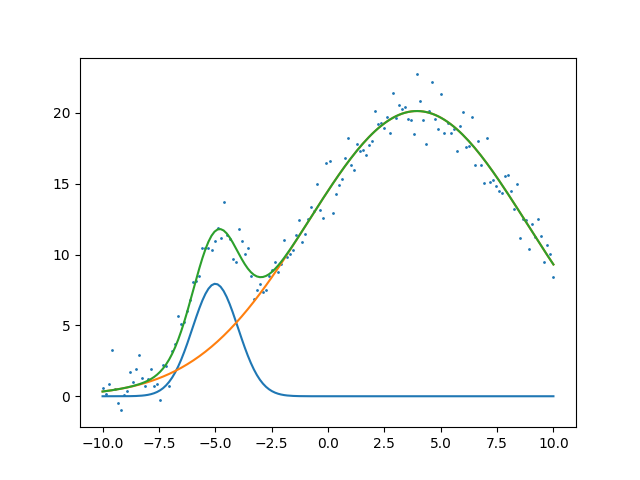
Now similarly we adapt our cost function for two gaussians and
optimize. We can see that this method does exactly what we would like
to do for our XRD data! I think at this point it is important to talk
about our initial guess. The initial guess is very important. The
computer science phrase garbage in, garbage out is extremely
relevant. If you play around with the initial guess in the code below
it will not always get the correct solution. This indicates that even
for this problem there are many local minimums. Try setting
initial_guess = np.random.randn(6) you will notice that it will get
the correct solution about one out of every ten times. This is where
domain knowledge comes in! Some things to know about profiles. The
amplitude is always positive and never exceeds the max amplitude in
the profile, the width is always positive and not extremely broad, and
the mean is always between the domain. With this information we now
have constraints that we can use to put bounds on our solution and
initial guess. We will use this information with lmfit.
|
|
steps 71 1.0378296281115338
g_0: amplitude: 20.102 mean: -4.999 sigma: 1.010
g_1: amplitude: 245.723 mean: 3.947 sigma: 4.871
And the resulting fit shown below.
|
|
At this point I think it is time that we try to fit actual XRD
data. We understand the method of fitting a profile. But the current
method of constructing the optimization is tedious to say the
least. As Raymond Hettinger always says there must be a better way!
Let’s use a package designed exactly for this lmfit. You can easily
install it via pip install lmfit. I think of lmfit as constructing a
model that represents your data by adding together several
functions. In our case these functions will be the gaussians,
lorentzians, and voigt. But lmfit is much more flexible than that see
the available functions and you can even include your own as an
arbitrary python function. Another issue that lmfit solves is mapping
your function parameters to the optimization routine and adding
complex constraints such as min, max, relationships between
parameters, and fixed values. Interestingly I found myself
constructing a similar class to their Parameter class in my code
DFTFIT.
|
|
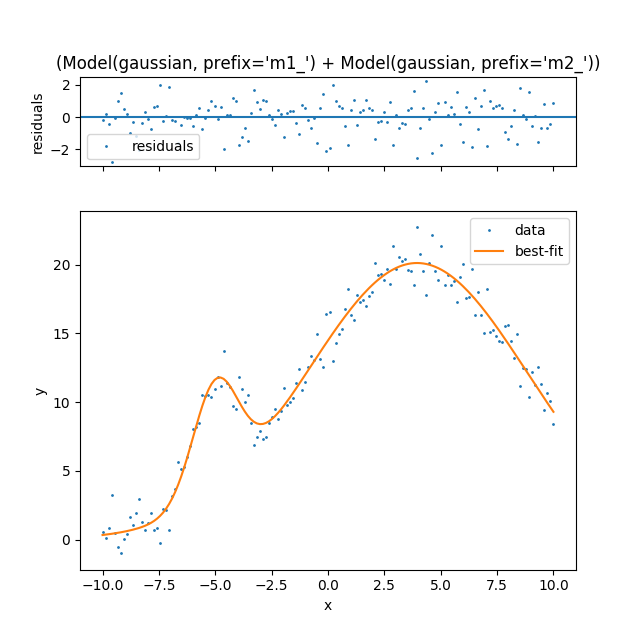
Notice how lmfit made the model construction much easier. You simply
add together your models and set the initial guess parameters for each
through make_params. If you would like to set bounds and additional
constraints use set_param_hint. Using the domain knowledge that we
have about XRD we can simplify the creation of these models
greatly. This will setup default initial guesses, set bounds on
variable values, and allow for a python dictionary as a spec.
|
|
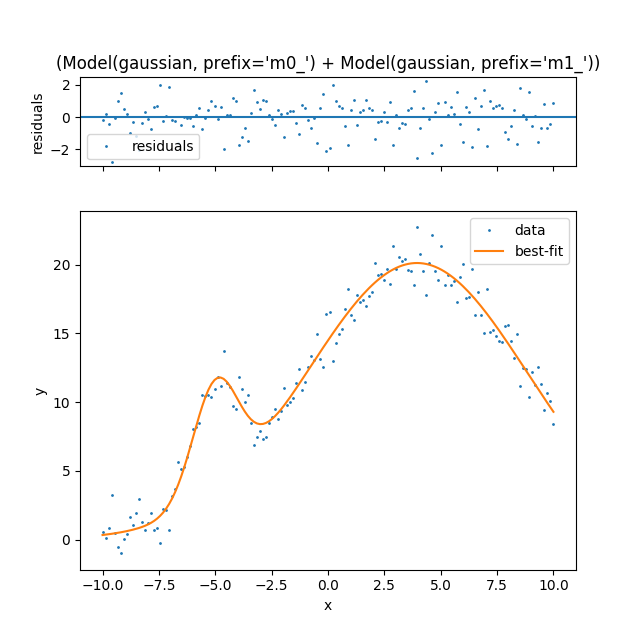
Lets try the same problem that we have been working on. Again to show
how there are multiple local optimums you will notice that this method
does not always converge to the true solution. The spec that we have
defined in the code above will work for gaussians, lorentzians, and
voigt. All parameters can be initialized via params and all hints
can be supplied as hints. I will show that in the final example.
|
|
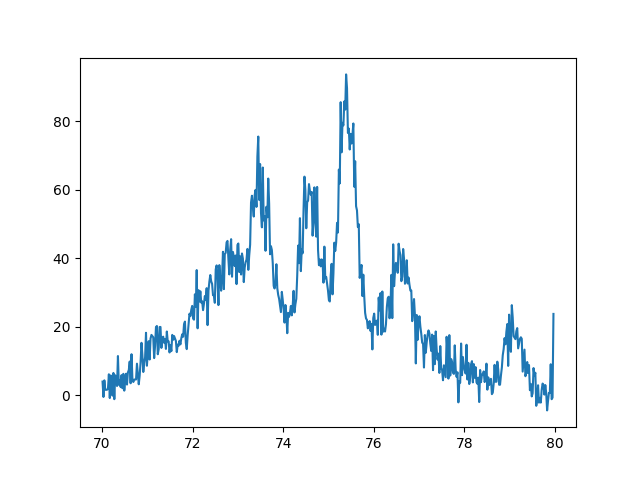
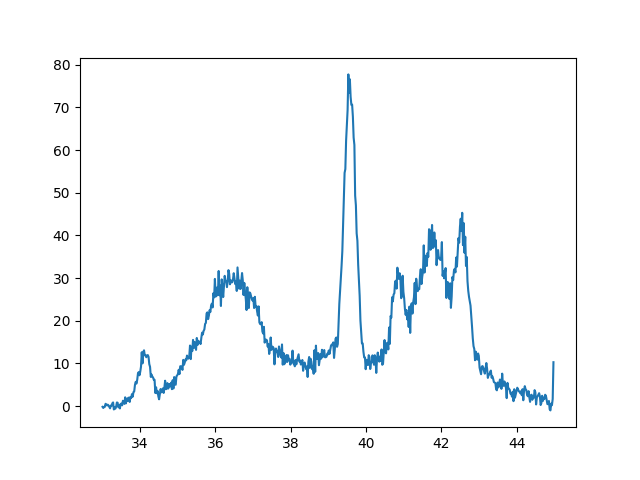
Finally here is our example XRD spectra. I will not show the actual data since my colleague would like to keep that for publication. But again here is the plot we are trying to fit. The data if from a very disordered sample due to irradiation.
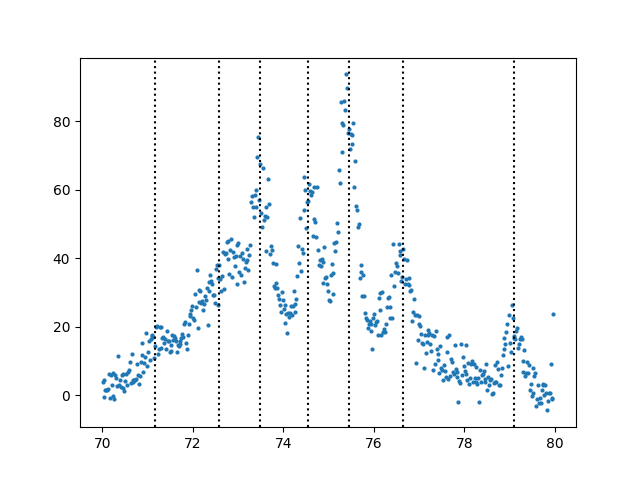
It is tedious to find all the peaks so lets write a function to help
us assign initial values for guesses based on peaks. This routine
uses scipy’s find_peaks_cwt method. I have found it to be superior
to many other peak finding algorithms out there.
|
|
Now using are good initial guesses for the peaks we begin our optimization.
|
|
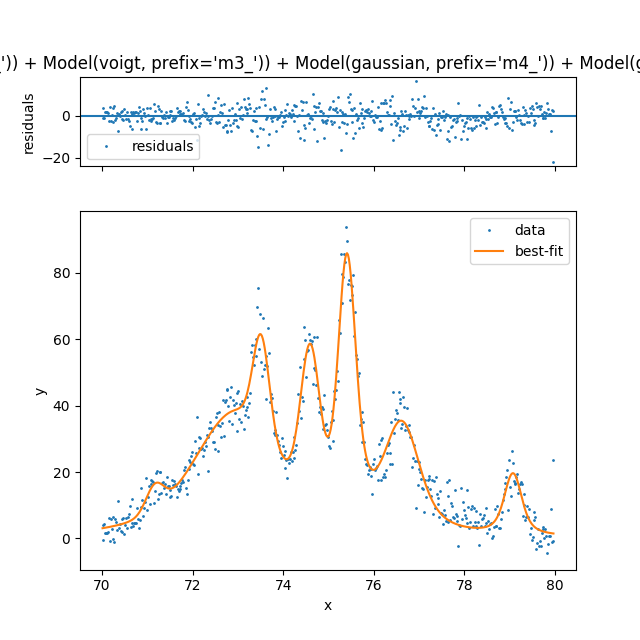
|
|
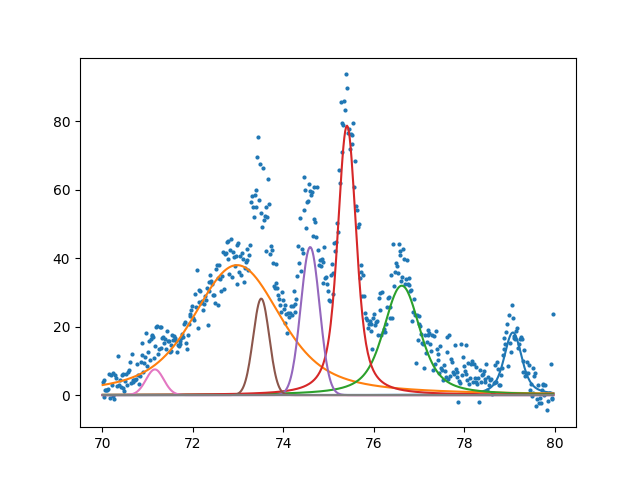
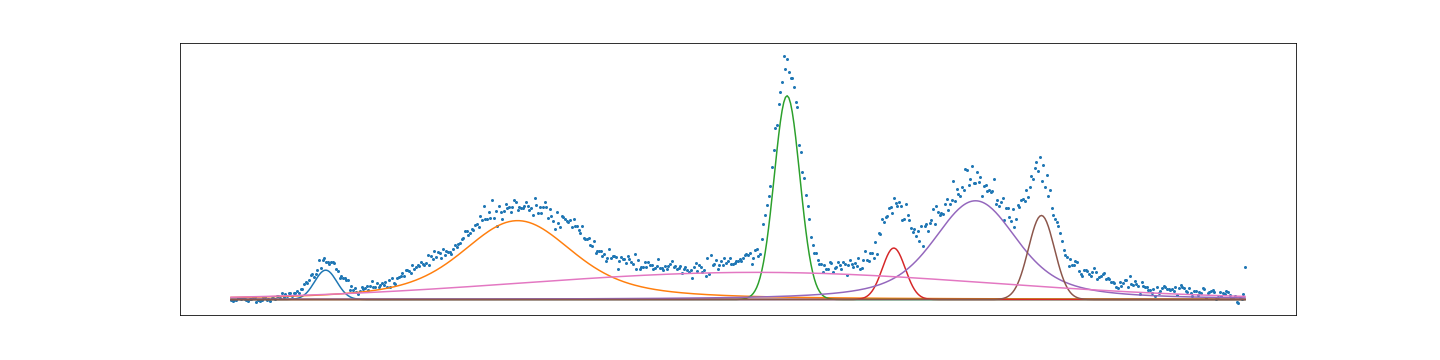
Finally showing that with a little bit of code we can automate a method for fitting that is equally impressive to commercial software. I would argue that this method is more flexible and does not hide the detail of fitting. Having a spec also makes it easy to save a model as a template for future fit. Something that I haven’t seen before. To show how detailed the spec can be here is an example of fitting a profile that has hidden peaks.
Here is the complex spec showing off all of the features. We see that it is possible to tightly constrain the location of peaks. Something that is not easily possible in other software.
|
|
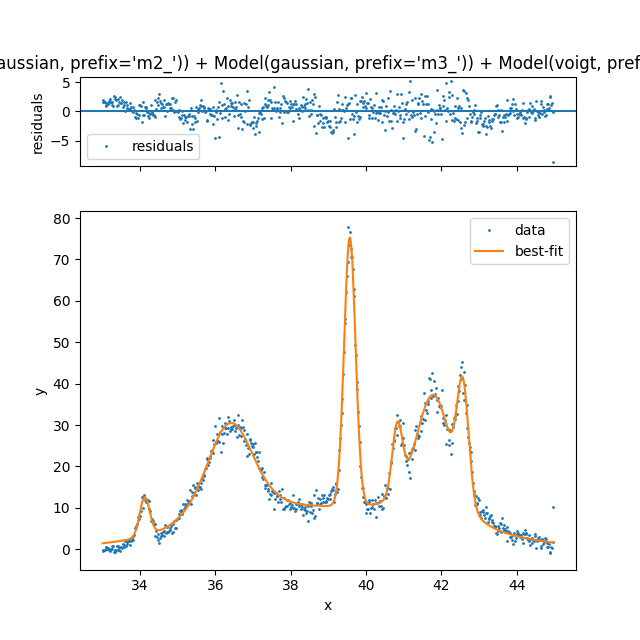
|
|
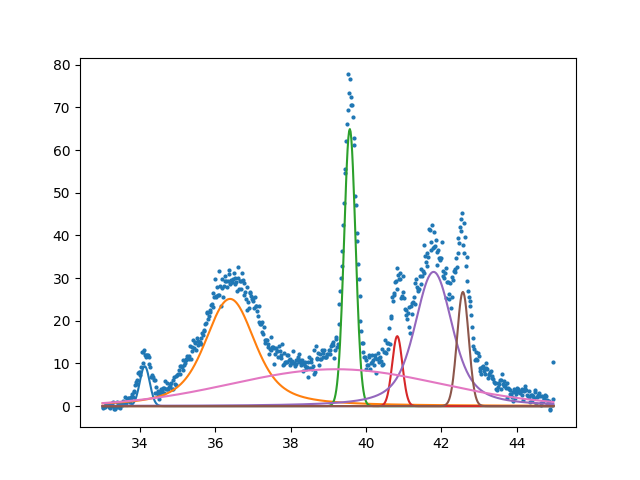
So far I have shown the fitting but we most importantly want the parameters for further analysis.
|
|
center model amplitude sigma gamma
[34.127] GaussianModel : 3.075, 0.131
[36.390] VoigtModel : 52.155, 0.433, 0.433
[39.568] GaussianModel : 24.495, 0.151
[40.827] GaussianModel : 5.531, 0.134
[41.791] VoigtModel : 49.129, 0.326, 0.326
[42.569] GaussianModel : 10.287, 0.153
[39.200] GaussianModel : 60.034, 2.769
I hope that this has been an informative tutorial on how powerful python with numpy, scipy, and lmfit is. All of the code was executed in an org babel environment so I can guarantee that the code should run in python 3.6. As always please leave a comment if I made an error or additional clarification is needed!